Direct Selection
To
be able to select for a cloned gene it is necessary
to plate the transformants onto an agar medium on which
only the desired recombinants, and no others, can grow.
The only colonies that are obtained will therefore be
ones that comprise cells containing the desired recombinant
DNA molecule.
The simplest example of Direct Selection occurs when
the desired gene specifies resistance to an antibiotic.
As an example will will consider an experiment to clone
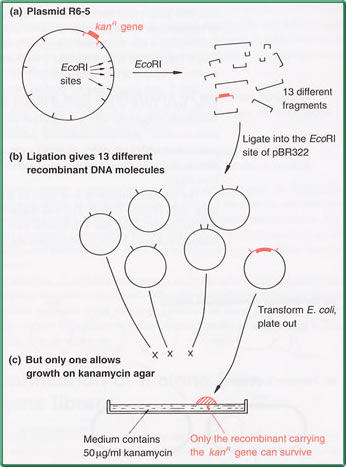
the gene for kanamcycin resistance from plasmid R6-5.
This plasmid carries genes for resistances to four antibiotics:
kanamycin, chloramphenicol, streptomycin and sulphonamide.
The kanamycin resistance gene lies within one of the
13 EcoRI fragments (a).
To clone this gene the EcoRI fragments of R6-5 would
be inserted into the EcoRI site of a vector such as
pBR322. The ligated mix will comprise many copies of
13 different recombinant DNA molecules, one set of which
carries the gene for kanamycin resistance.
Insertional inactivation cannot be used to select recombinants
when the EcoRI site of pBR322 is used. This is becuase
this site does not lie in either the ampicillin or the
tetracycline resistance of this plasmid.
But this is
immaterial for cloning the kanamycin resistance gene
because in this case the coloned gene can be used as
the selectable marker. Transformants are plated onto
kanamycin agar, on which the only cells able to survice
and produce colonies are those recombinants that contain
the cloned kanamycin resistance gene(c).
Next: Extending
the Scope of Direct Selection...
|

